Laboratory of Regulatory RNAs
Head:
Where to Find Us:
Viničná 5, Prague 2 - New Town, 128 43
the ground floor (room 006)
Team MembersMethodology and Technical SupportScientific CollaborationsPublications |
TeachingProposed Topics of Theses for New StudentsStudent Theses in ProgressPast Student Theses |
Research Interests
Further information can be also found at the https://web.natur.cuni.cz/molbio/hnilicovalab/ website
Laboratory of Regulatory RNAs was established at the end of 2023 as the newest research group of the Department of Genetics and Microbiology, Faculty of Science, Charles University. Prior to this, it was part of the Laboratory of Microbial Genetics and Gene Expression at the Institute of Microbiology of the Czech Academy of Sciences. The group focuses on small non-coding RNAs that regulate bacterial transcription. Our primary interest lies in RNAs that bind to RNA polymerases (RNAP). Our long-term goal is to understand how these RNAs function and to explore whether they could be utilized in the future for the development of specific RNA antibiotics targeting individual bacterial species.
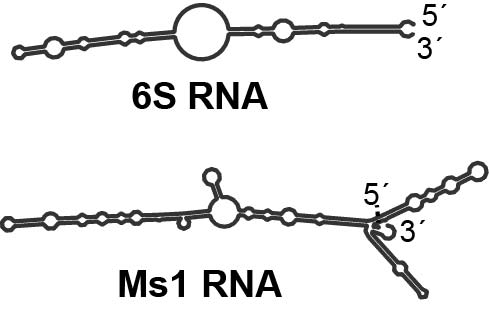 As early as 2013, we discovered that Ms1 RNA, which is present in significant amounts during the stationary phase of the bacterial life cycle in Mycobacterium smegmatis, can bind to the core RNAP proteins without the help of sigma factors (only the second RNA known to do this, after 6S RNA, which is not present in M. smegmatis). Additionally, we demonstrated that Ms1 RNA regulates the amount of RNAP in this bacterium.
As early as 2013, we discovered that Ms1 RNA, which is present in significant amounts during the stationary phase of the bacterial life cycle in Mycobacterium smegmatis, can bind to the core RNAP proteins without the help of sigma factors (only the second RNA known to do this, after 6S RNA, which is not present in M. smegmatis). Additionally, we demonstrated that Ms1 RNA regulates the amount of RNAP in this bacterium.
The Laboratory uses the native RIP-seq method to detect RNAs interacting with RNAP in various bacteria. In collaboration with bioinformaticians, we can distinguish between newly emerging transcripts associated with transcribing RNAP, and RNAs that bind to non-transcribing RNAP. In collaboration with the Ministry of Defense of the Czech Republic, we performed RIP-seq with M. tuberculosis in their BSL3 laboratory, and we showed that the homolog of Ms1, named MTS2823, also interacts with RNAP during the stationary phase of M. tuberculosis life cycle. The M. smegmatis RIP-seq and M. smegmatis NGS data are now publicly available at http://msmegseq.elixir-czech.cz/ website.
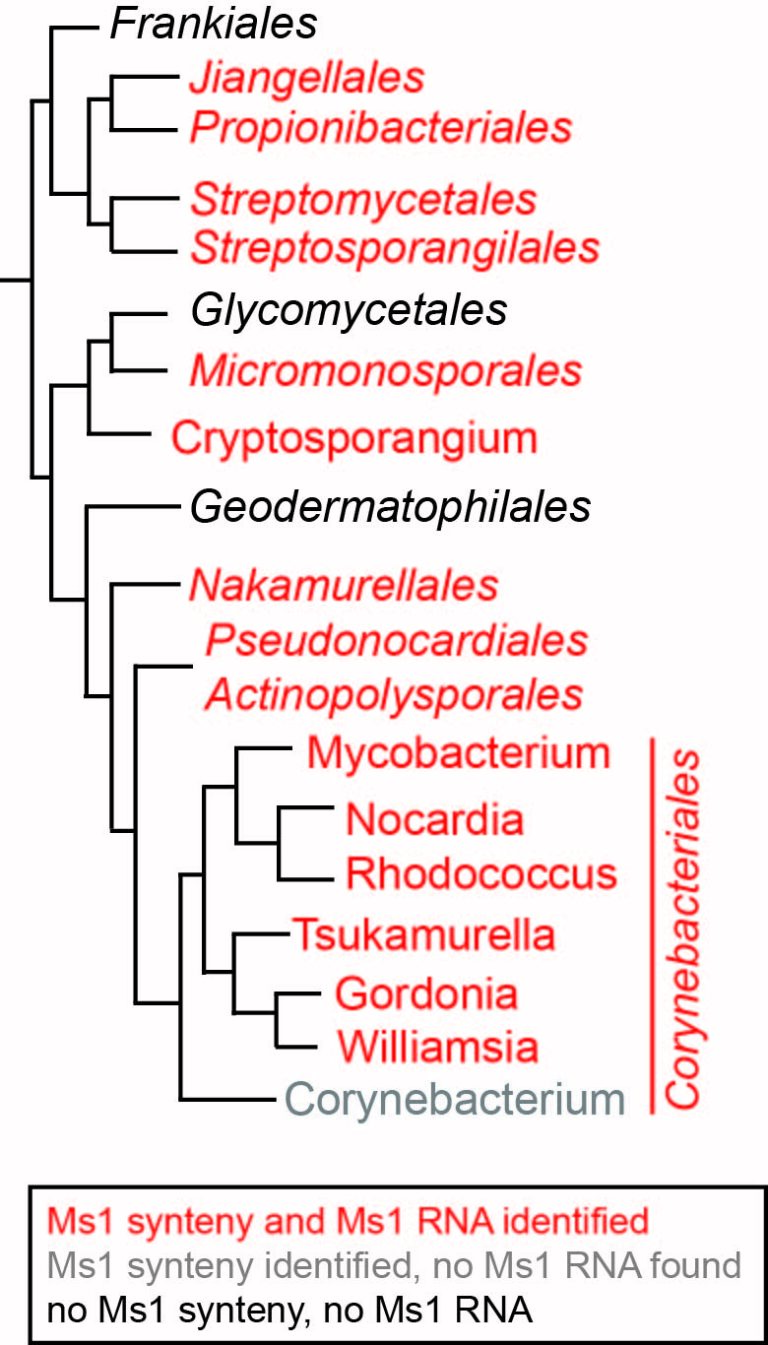 Another question was whether Ms1 RNA is present in bacteria other than Mycobacterium species. With the help of bioinformatics approaches, we successfully identified Ms1 homologs in various actinobacteria and in Streptomyces coelicolor. Surprisingly, however, in several bacterial species (e.g., Corynebacterium glutamicum), we did not detect the presence of either 6S RNA or Ms1 RNA. Nevertheless, using RIP-seq in C. glutamicum, we discovered a new RNA, approximately 500 nucleotides long, capable of binding to both forms of RNAP: RNAP core and RNAP holoenzyme (with the primary sigma factor). This RNA has a unique secondary structure and length, unlike 6S and Ms1 RNA, making it undetectable by bioinformatics approaches alone. It is possible that other bacteria lacking 6S/Ms1/CoRP RNA may have additional new types of RNAP-binding RNAs. Identifying these other RNAs will contribute to our understanding of the variability of RNA structures capable of associating with RNAP.
Another question was whether Ms1 RNA is present in bacteria other than Mycobacterium species. With the help of bioinformatics approaches, we successfully identified Ms1 homologs in various actinobacteria and in Streptomyces coelicolor. Surprisingly, however, in several bacterial species (e.g., Corynebacterium glutamicum), we did not detect the presence of either 6S RNA or Ms1 RNA. Nevertheless, using RIP-seq in C. glutamicum, we discovered a new RNA, approximately 500 nucleotides long, capable of binding to both forms of RNAP: RNAP core and RNAP holoenzyme (with the primary sigma factor). This RNA has a unique secondary structure and length, unlike 6S and Ms1 RNA, making it undetectable by bioinformatics approaches alone. It is possible that other bacteria lacking 6S/Ms1/CoRP RNA may have additional new types of RNAP-binding RNAs. Identifying these other RNAs will contribute to our understanding of the variability of RNA structures capable of associating with RNAP.





















