Laboratory of RNA Biochemistry
Head:
RNDr. Martin Pospíšek, Ph.D.
Department of Genetics and Microbiology
Study consultant and advisor for M.Sc. specialization Molecular Biology and Genetics of Eukaryotes
Viničná 5, 2NP, room 114
Where to Find Us:
Viničná 5, Prague 2 - New Town, 128 43
the ground floor (room P10), the basement (rooms S12, S14), the first floor (rooms 112, 114, 115)
Team MembersMethodology and Technical SupportScientific CollaborationsPublications |
TeachingProposed Topics of Theses for New StudentsStudent Theses in ProgressPast Student Theses |
Research Interests
Further information can be also found at http://web.natur.cuni.cz/~pospisek website
 The Laboratory of RNA Biochemistry primarily focuses on studying eukaryotic protein synthesis and its relationship to gene expression regulation. Our research combines bioinformatics and experimental approaches. We have created and maintain two publicly accessible databases: http://iresite.org/ and http:/hcvivdb.org. In 2024, as part of the "Pathogen-Host Interactions" and "Immunity Against Viruses and Bacteria" programmes, we became part of the National Institute of Virology and Bacteriology, a platform that brings together leading scientific institutions from the whole Czech Republic.
The Laboratory of RNA Biochemistry primarily focuses on studying eukaryotic protein synthesis and its relationship to gene expression regulation. Our research combines bioinformatics and experimental approaches. We have created and maintain two publicly accessible databases: http://iresite.org/ and http:/hcvivdb.org. In 2024, as part of the "Pathogen-Host Interactions" and "Immunity Against Viruses and Bacteria" programmes, we became part of the National Institute of Virology and Bacteriology, a platform that brings together leading scientific institutions from the whole Czech Republic.
Our research interests can be divided into three main areas:
1) Cap-dependent translation initiation - unexpected functions of the eIF4E translation initiation factors
Protein synthesis is the primary energy expenditure for the cell, so it is not surprising that this process is closely monitored and regulated from the very beginning. Cellular mRNA carries a cap (m7GTP) at its 5'-end, which is necessary for recognition by the translation initiation factor eIF4E1, subsequently facilitating the binding to the cellular translation machinery.
In higher eukaryotes, paralogous genes encoding the eIF4E family of translation factors have evolved. These proteins, although very similar to the canonical eIF4E1 at the sequence level, have acquired special properties, such as regulating the preferential translation of certain mRNAs, mediating translational inhibition, or having tissue- or development-specific expression. In the human genome, besides the gene for the prototype eIF4E1, there are also genes for eIF4E2 (4EHP, class II of the protein family) and eIF4E3 (class III).
 We performed a bioinformatic analysis of the human 4E family genes and found that each gene produces several biologically significant, sequence-divergent mRNAs. We mapped the 3'-ends of these transcripts in detail and experimentally confirmed the use of multiple polyadenylation sites. We obtained cDNA of selected variants and used them to create N-terminal fusions with the GFP coding sequence. This allowed us to create stable cell lines with inducible expression of specific 4E proteins tagged with a green fluorescent marker. This system was used to describe the cellular localization of individual 4E proteins, leading to the surprising finding that each of the studied proteins exhibits a unique localization pattern regarding its presence in P-bodies and stress granules. This result also indicates the unique and indispensable function of each of these initiation factors in the cell. Our current interest lies in identifying previously unknown interaction partners of the eIF4E2 and eIF4E3 proteins, and in finding cellular transcripts specifically bound by these two initiation factors. The research on eIF4E1 and eIF4E2 factors is complemented by studies in the beetle Tribolium castaneum, focusing on developmental defects after silencing the respective genes.
We performed a bioinformatic analysis of the human 4E family genes and found that each gene produces several biologically significant, sequence-divergent mRNAs. We mapped the 3'-ends of these transcripts in detail and experimentally confirmed the use of multiple polyadenylation sites. We obtained cDNA of selected variants and used them to create N-terminal fusions with the GFP coding sequence. This allowed us to create stable cell lines with inducible expression of specific 4E proteins tagged with a green fluorescent marker. This system was used to describe the cellular localization of individual 4E proteins, leading to the surprising finding that each of the studied proteins exhibits a unique localization pattern regarding its presence in P-bodies and stress granules. This result also indicates the unique and indispensable function of each of these initiation factors in the cell. Our current interest lies in identifying previously unknown interaction partners of the eIF4E2 and eIF4E3 proteins, and in finding cellular transcripts specifically bound by these two initiation factors. The research on eIF4E1 and eIF4E2 factors is complemented by studies in the beetle Tribolium castaneum, focusing on developmental defects after silencing the respective genes.
2) Cap-independent translation, HCV IRES, and pGKL plasmids
Some viral and cellular RNAs are translated independently of the cap by directly binding to ribosomes using so-called IRES elements (Internal Ribosome Entry Sites). Therefore, another focus of our research is the Hepatitis C Virus (HCV). The ability to produce viral proteins is a prerequisite for productive infection, which in this case is fully dependent on a functional IRES element. Finding substances that inhibit IRES function could lead to the introduction of an effective treatment for this currently incurable disease. The serious health concerns are highlighted by the World Health Organization, which labels HCV infection as a "ticking bomb", as around 200 million people worldwide are infected with this virus.
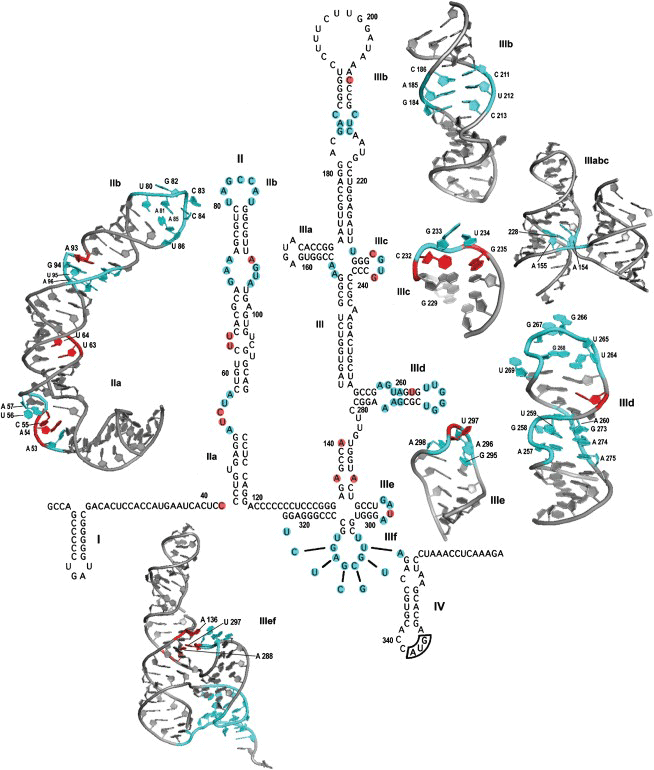 Our interest in IRES-mediated translation led to the creation of the IRESite database IRESite (http://iresite.org/), which contains available data on experimentally verified IRES sequences and structures and is currently the only maintained database of its kind. We also focused on methodically processing available information about the impact of primary IRES sequence changes on its secondary structure and the functionality of its individual domains. In connection with this, we launched the HCVIVdb database HCVIVdb (HCV IRES Variation Database; http:/hcvivdb.org). Recently, we established a model system in our Laboratory that allows studying IRES directly in yeast cells. We are also optimizing the use of a bicistronic reporter system for screening libraries of biologically active substances to find an HCV IRES translation inhibitor.
Our interest in IRES-mediated translation led to the creation of the IRESite database IRESite (http://iresite.org/), which contains available data on experimentally verified IRES sequences and structures and is currently the only maintained database of its kind. We also focused on methodically processing available information about the impact of primary IRES sequence changes on its secondary structure and the functionality of its individual domains. In connection with this, we launched the HCVIVdb database HCVIVdb (HCV IRES Variation Database; http:/hcvivdb.org). Recently, we established a model system in our Laboratory that allows studying IRES directly in yeast cells. We are also optimizing the use of a bicistronic reporter system for screening libraries of biologically active substances to find an HCV IRES translation inhibitor.
Another topic of our research is the post-transcriptional and translational regulation of transcripts originating from pGKL plasmids. Plasmids pGKL1 and 2 are naturally found in the cytoplasm of the yeast Kluyveromyces lactis. Fifteen annotated open reading frames encode experimentally verified or putative components of the transcriptional apparatus, including some enzymes mediating cap synthesis. We are currently studying the mechanism by which the 5'- and 3'-ends of plasmid transcripts are formed and how their translation proceeds.
3) Study of protein-protein interactions and nuclear functions of Interleukin-1α using yeast and human tissue cultures
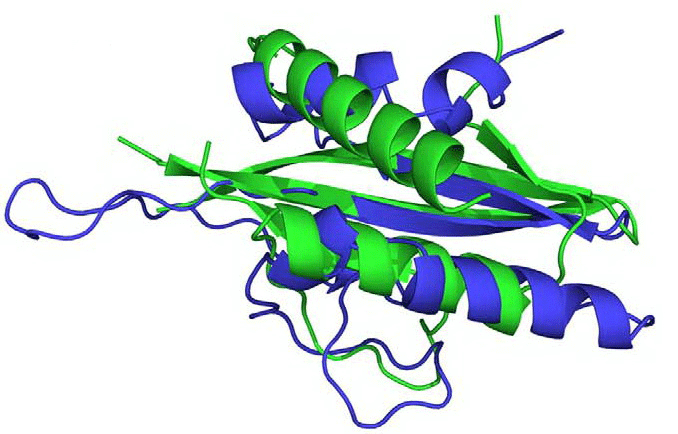 The pro-inflammatory Interleukin-1α (Il-1α) is a key player in immune responses in higher eukaryotes. Although its effects on different cell types are quite diverse, it has been shown to be associated with a range of autoimmune and cardiovascular diseases, including rheumatoid arthritis and Alzheimer’s disease. Surprisingly, a significant portion of Il-1α translocates to the nucleus, where it interacts with histone acetyltransferase complexes. In our previous work, using immunoprecipitation and yeast deletion strains, we identified the binding site for the Il-1α precursor in the HAT/Core module of the yeast SAGA complex. We also discovered a surprising structural similarity between the N-terminal domain of Il-1α and the C-regulatory domain of the yeast AMP-activated protein kinase Snf1, which interacts with the HAT complex. This finding is particularly exciting given that the same interaction has been described for the human homolog of Snf1. This discovery is a continuation of our long-standing interest in cytokine signaling and related transcriptional regulation.
The pro-inflammatory Interleukin-1α (Il-1α) is a key player in immune responses in higher eukaryotes. Although its effects on different cell types are quite diverse, it has been shown to be associated with a range of autoimmune and cardiovascular diseases, including rheumatoid arthritis and Alzheimer’s disease. Surprisingly, a significant portion of Il-1α translocates to the nucleus, where it interacts with histone acetyltransferase complexes. In our previous work, using immunoprecipitation and yeast deletion strains, we identified the binding site for the Il-1α precursor in the HAT/Core module of the yeast SAGA complex. We also discovered a surprising structural similarity between the N-terminal domain of Il-1α and the C-regulatory domain of the yeast AMP-activated protein kinase Snf1, which interacts with the HAT complex. This finding is particularly exciting given that the same interaction has been described for the human homolog of Snf1. This discovery is a continuation of our long-standing interest in cytokine signaling and related transcriptional regulation.
Methodological research and other activities
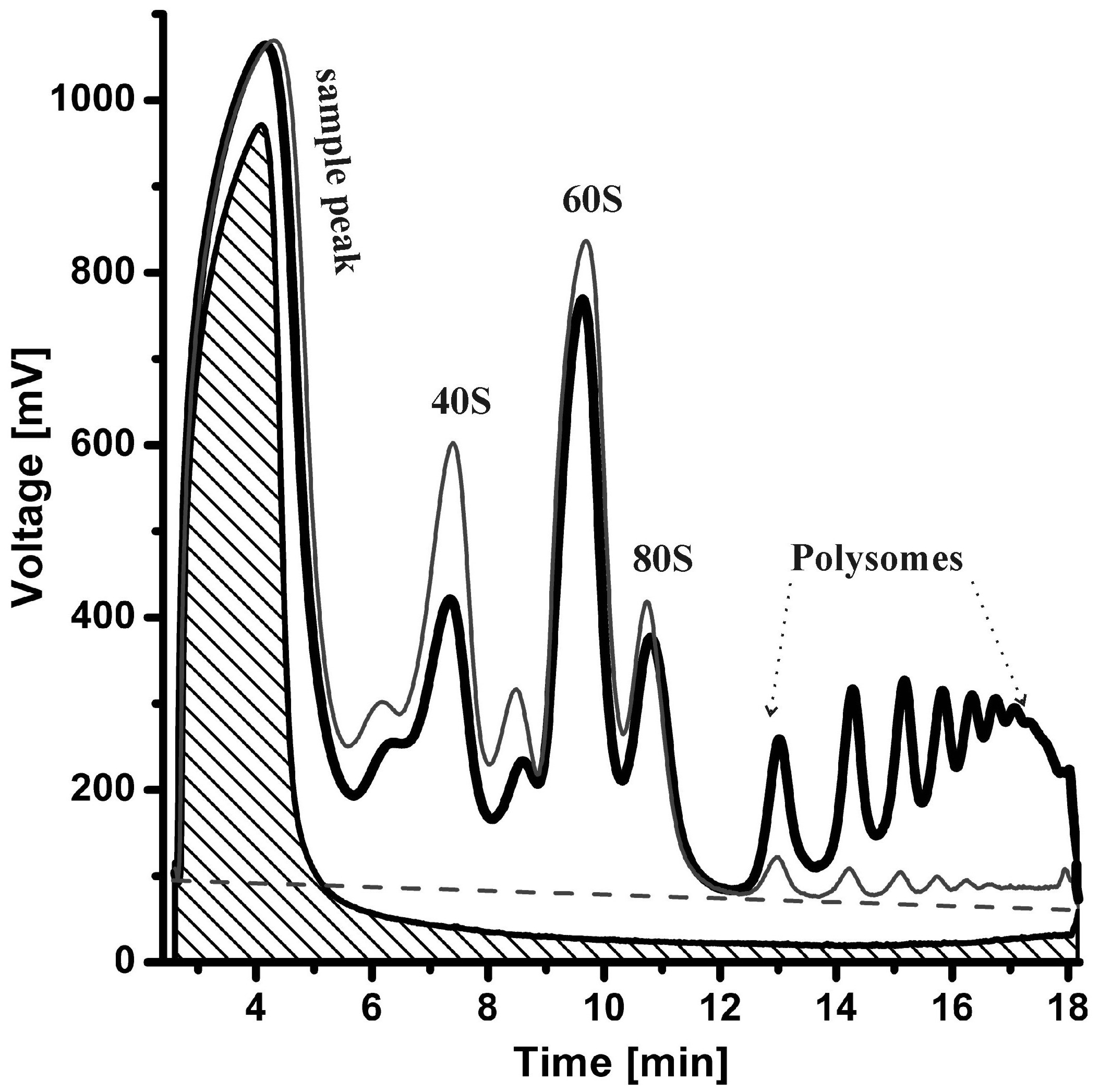 During our research on cap-independent translation, we published several methodological papers. We described cryptic transcription in the widely used luciferase reporter gene, which will help eliminate some methodological issues in IRES characterization in the future. In 2005, we published a simplified protocol for RNA electrophoresis in agarose gels. This article has been one of the most downloaded publications in Analytical Biochemistry for several years and was awarded by Elsevier. We contributed to the improvement of the polysome profiling method, which we described in two review articles.
During our research on cap-independent translation, we published several methodological papers. We described cryptic transcription in the widely used luciferase reporter gene, which will help eliminate some methodological issues in IRES characterization in the future. In 2005, we published a simplified protocol for RNA electrophoresis in agarose gels. This article has been one of the most downloaded publications in Analytical Biochemistry for several years and was awarded by Elsevier. We contributed to the improvement of the polysome profiling method, which we described in two review articles.
The Laboratory team is also active in organizing scientific life. In 2003, our Laboratory founded the tradition of annual RNA Clubs, many of which were also organized by our members, and these have become a regular meeting place for many researchers interested in RNA topics.





















