Laboratory of Bacterial Genetics
Head:
RNDr. Jaroslav Nunvář, Ph.D.
Where to Find Us:
Viničná 5, Prague 2 - New Town, 128 43
the first floor (room 110)
Team MembersMethodology and Technical SupportScientific CollaborationsPublications |
TeachingProposed Topics of Theses for New StudentsStudent Theses in ProgressPast Student Theses |
Research Interests
 The Laboratory of Bacterial Genetics was established in 2010 under the leadership of Irena Lichá, officially branching off from the Laboratory of Bacterial Physiology (although even before there was some research done in this field by Josef Náprstek, Ph.D.). This reflected the focus of its early research on mechanisms that allow soil bacteria to cope with various stressors and survive in their natural environments. Soil bacteria are highly adept at adjusting to rapidly changing conditions, such as daily temperature fluctuations or shifts in osmolarity due to soil drying and rehydration. Moreover, they constantly engage in competition for territory and resources. Bacteria can adapt to these conditions not only by altering their overall life strategies through the "turning on" and "off" the expression of specific genes but may also benefit from horizontal acquisition of fitness-enhancing genes. Within the scope of research on the effects of physical factors on bacterial cells, the laboratory primarily studied the molecular basis of the regulation of the so-called general stress response, using the soil bacterium Bacillus subtilis as its model.
The Laboratory of Bacterial Genetics was established in 2010 under the leadership of Irena Lichá, officially branching off from the Laboratory of Bacterial Physiology (although even before there was some research done in this field by Josef Náprstek, Ph.D.). This reflected the focus of its early research on mechanisms that allow soil bacteria to cope with various stressors and survive in their natural environments. Soil bacteria are highly adept at adjusting to rapidly changing conditions, such as daily temperature fluctuations or shifts in osmolarity due to soil drying and rehydration. Moreover, they constantly engage in competition for territory and resources. Bacteria can adapt to these conditions not only by altering their overall life strategies through the "turning on" and "off" the expression of specific genes but may also benefit from horizontal acquisition of fitness-enhancing genes. Within the scope of research on the effects of physical factors on bacterial cells, the laboratory primarily studied the molecular basis of the regulation of the so-called general stress response, using the soil bacterium Bacillus subtilis as its model.
In the following years, the research focus shifted to bacterial persistence, i.e., the ability of bacteria to survive antibiotic exposure without developing genetically based, hereditary resistance. This phenomenon is closely related to stress adaptations and complicates the antimicrobial treatment of chronically ill patients. Persistence was studied in the bacterium Staphylococcus aureus, a distant relative to the model organism B. subtilis and a key opportunistic pathogen in immunocompromised patients. An important part of the research was a comparative analysis of whole genomes of clonally related S. aureus isolated from individual patients during chronic infection. Similar analysis was also conducted for an epidemic clone of Burkholderia cenocepacia, which causes fatal chronic infections in Czech patients with cystic fibrosis, and for the so-called "new European clone" of enterohemorrhagic Escherichia coli (EHEC).
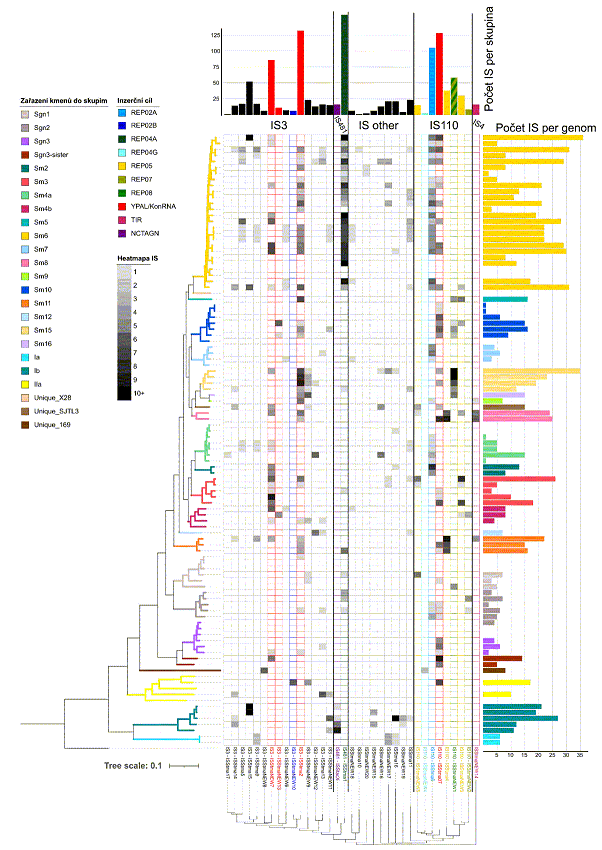
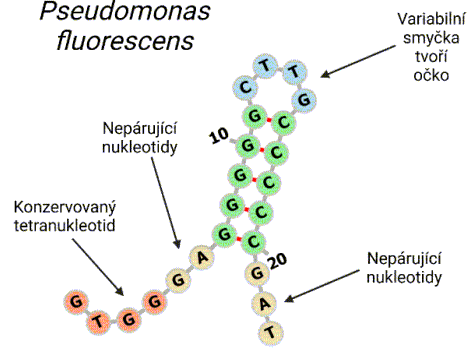 Currently, under the leadership of Jaroslav Nunvář, Ph.D., the Laboratory focuses mainly on bioinformatic analysis of repetitive sequences in bacterial genomes. This research direction follows an original "in house" discovery made in the Laboratory in 2009, which, based on analysis of Stenotrophomonas genomes, predicted a functional
Currently, under the leadership of Jaroslav Nunvář, Ph.D., the Laboratory focuses mainly on bioinformatic analysis of repetitive sequences in bacterial genomes. This research direction follows an original "in house" discovery made in the Laboratory in 2009, which, based on analysis of Stenotrophomonas genomes, predicted a functional
connection between tyrosine nucleases (RAYT) and extragenic repetitive DNA sequences (REP elements). The ability of RAYT nucleases to mobilize REP elements was later confirmed through structural studies. The Laboratory's research has significantly expanded knowledge of the REP/RAYT system (taxonomical distribution, structural conservation/diversity). Our recent results indicate that REP elements are frequent targets of insertions by transposable elements (insertion sequences, "jumping genes"). Analysis of Stenotrophomonas genomes further yielded a new class of repetitive elements (YPAL-like), characterized by a strongly conserved secondary structure and moderately conserved sequence. Our research aims to gain a deepen understanding of the extragenic space of bacterial genomes, which is very dynamic and highly variable across bacterial taxa.





















