Focus of the Molecular Biology of Plants group
Other topics covered in our group |
|
|
Glutamine synthetase GS2 in A. thaliana and its role in photorespiration and primary nitrogen assimilation |
Genetic and epigenetic causes of auxin autonomy in tobacco cell lines |
|
PsbO protein isoforms and their role in the photochemical phase of photosynthesis in A. thaliana |
Regulator of G-protein signaling (RGS1) and its role in potato development |
Introduction to epigenetics
Genetic information remains almost identical in all cells of multicellular organisms throughout their development and also regardless of environmental conditions. Since various gene products are needed under various conditions, the activity of particular genome regions must be flexibly regulated. This is particularly true for plants, which tolerate a large number of transposable elements (TEs) interspersed between genes in their genomes, with TEs accounting for up to 85% of the genome size. Plants can also cope effectively with and benefit from the whole-genome duplications that drive their evolution. For the proper functioning of such genomes, plants have developed a wide range of epigenetic tools to mark chromatin at both the DNA and histone levels. Moreover, these epigenetic pathways are tightly interconnected, increasing the robustness of the whole system. Histone labelling is highly complex and involves a variety of post-translational modifications of histone tails or different representations of histone variants in nucleosomes. DNA is labelled almost exclusively by methylation of cytosines, which may have various roles depending on the sequence context in which they are found. Chromatin marks are often only transient or easily reversible but can also be very stable and transgenerationally inherited. Therefore, the de novo establishment of epigenetic marks must be well-founded and highly specific. Silencing of chromatin activity can be mediated by DNA methylation, which is de novo guided into active regions of the genome by small RNAs (sRNAs in complex with their effector proteins, Argonaute, AGO) in a process called "RNA-directed DNA methylation" (RdDM). RdDM, in which plant-specific RNA polymerase V plays a key role, thus links chromatin modifications to RNA interference (RNAi). RNAi regulates gene expression via sRNA at the post-transcriptional level. Therefore, small RNAs can silence gene expression at the transcriptional and post-transcriptional levels (transcriptional and post-transcriptional gene silencing, TGS/PTGS).
Regulation of gene expression and chromatin state by small RNAs and RNA polymerases II and V
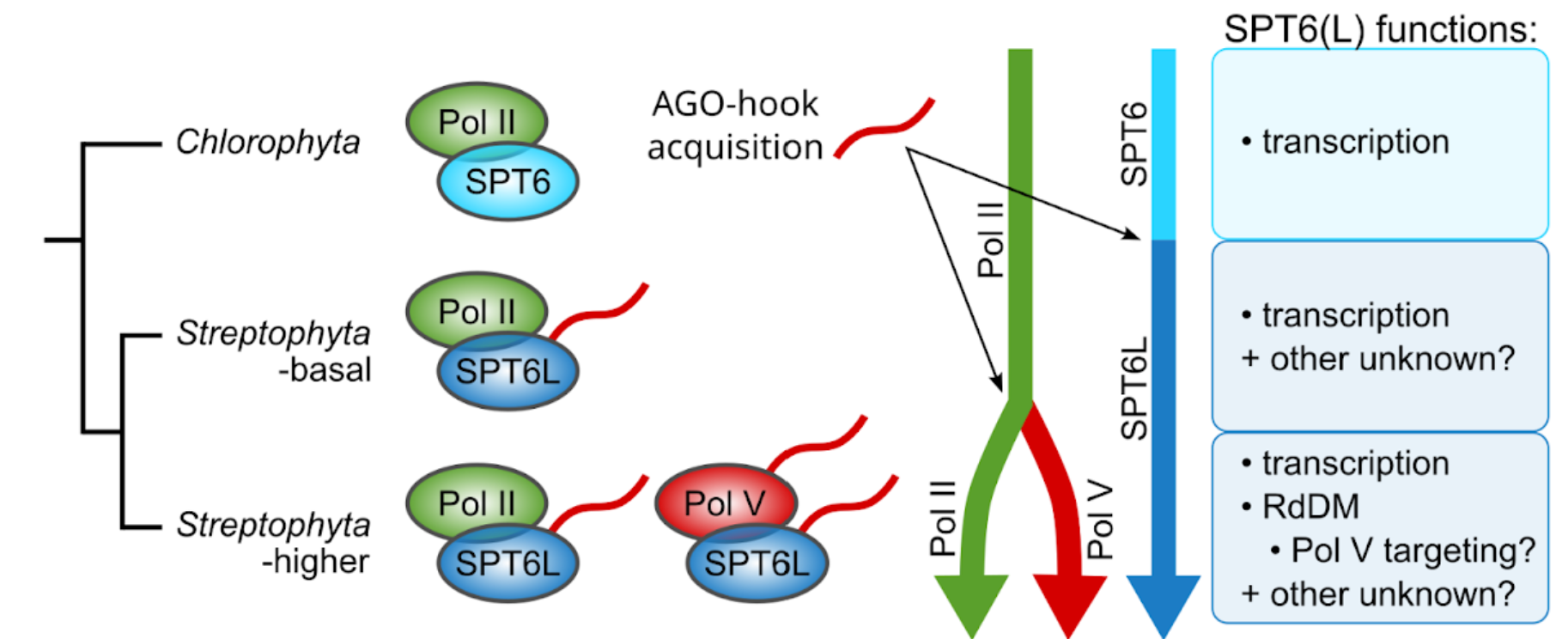 WHAT WE ALREADY KNOW? The mechanisms that ensure the maintenance of DNA methylation across cell divisions are relatively well described. The RdDM pathway is crucial for targeting DNA methylation to unmethylated regions of the genome. However, it is unclear how RdDM components are guided to the active chromatin to induce de novo methylation. The protein SPT6L could play a role in this process. It is a histone chaperone and transcription elongation factor of Pol II and V that can also participate in transcription initiation. In plants, SPT6L has an additional domain capable of binding AGO proteins, sRNA effectors. SPT6L acquired this domain at a very early stage of plant evolution before the RdDM pathway emerged. More details can be found in our paper 'SPT6L, a newly discovered ancestral component of the plant RNA-directed DNA methylation pathway'.
WHAT WE ALREADY KNOW? The mechanisms that ensure the maintenance of DNA methylation across cell divisions are relatively well described. The RdDM pathway is crucial for targeting DNA methylation to unmethylated regions of the genome. However, it is unclear how RdDM components are guided to the active chromatin to induce de novo methylation. The protein SPT6L could play a role in this process. It is a histone chaperone and transcription elongation factor of Pol II and V that can also participate in transcription initiation. In plants, SPT6L has an additional domain capable of binding AGO proteins, sRNA effectors. SPT6L acquired this domain at a very early stage of plant evolution before the RdDM pathway emerged. More details can be found in our paper 'SPT6L, a newly discovered ancestral component of the plant RNA-directed DNA methylation pathway'.
WHY ARE WE DOING THIS? We are interested in the significance of the AGO-binding domain: what role does it play in the Pol II complex? What role it has in the Pol V complex, where several other proteins have a similar AGO-binding domain? Could it be involved in guiding Pol V to unmethylated stretches of DNA?
DNA repair and CRISPR/Cas mutagenesis and how they are affected by small RNAs and chromatin state
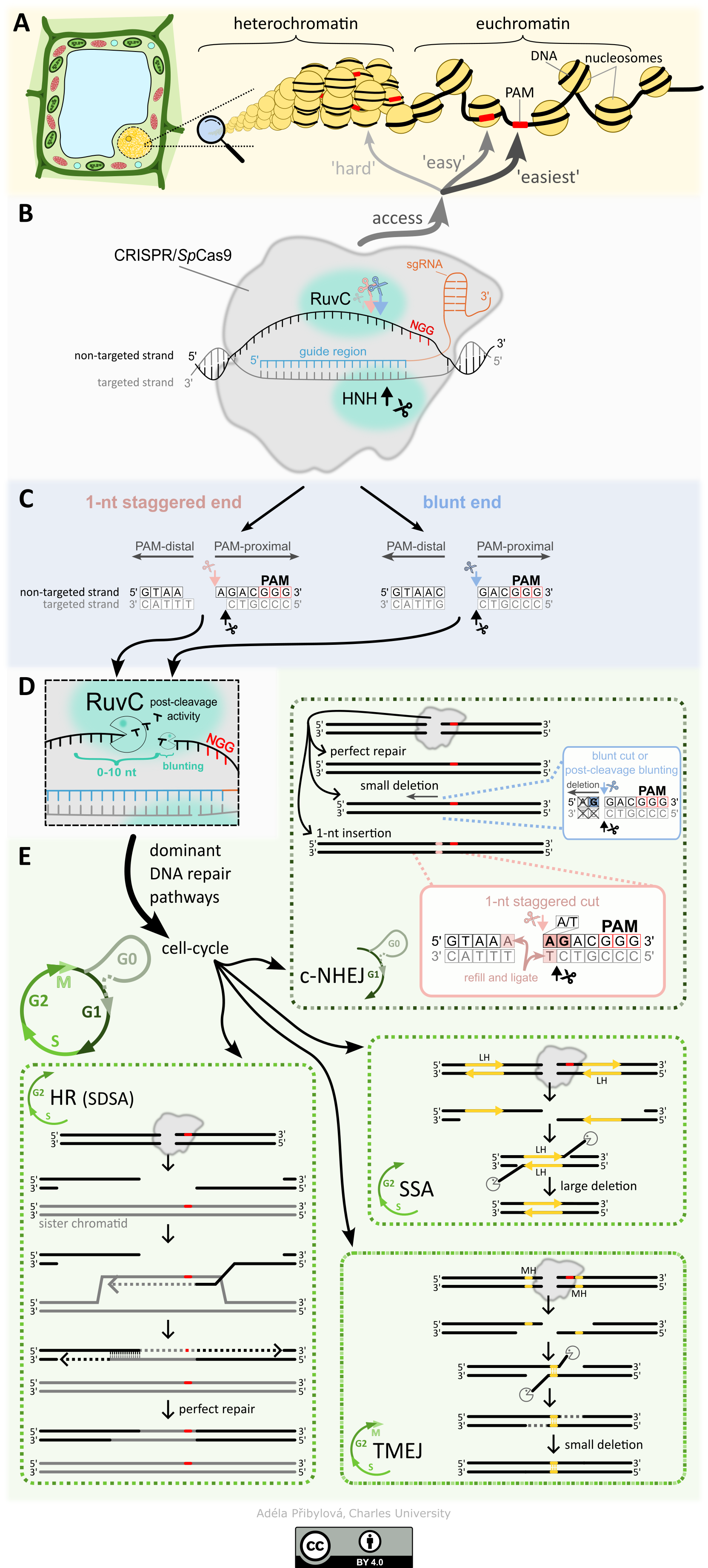
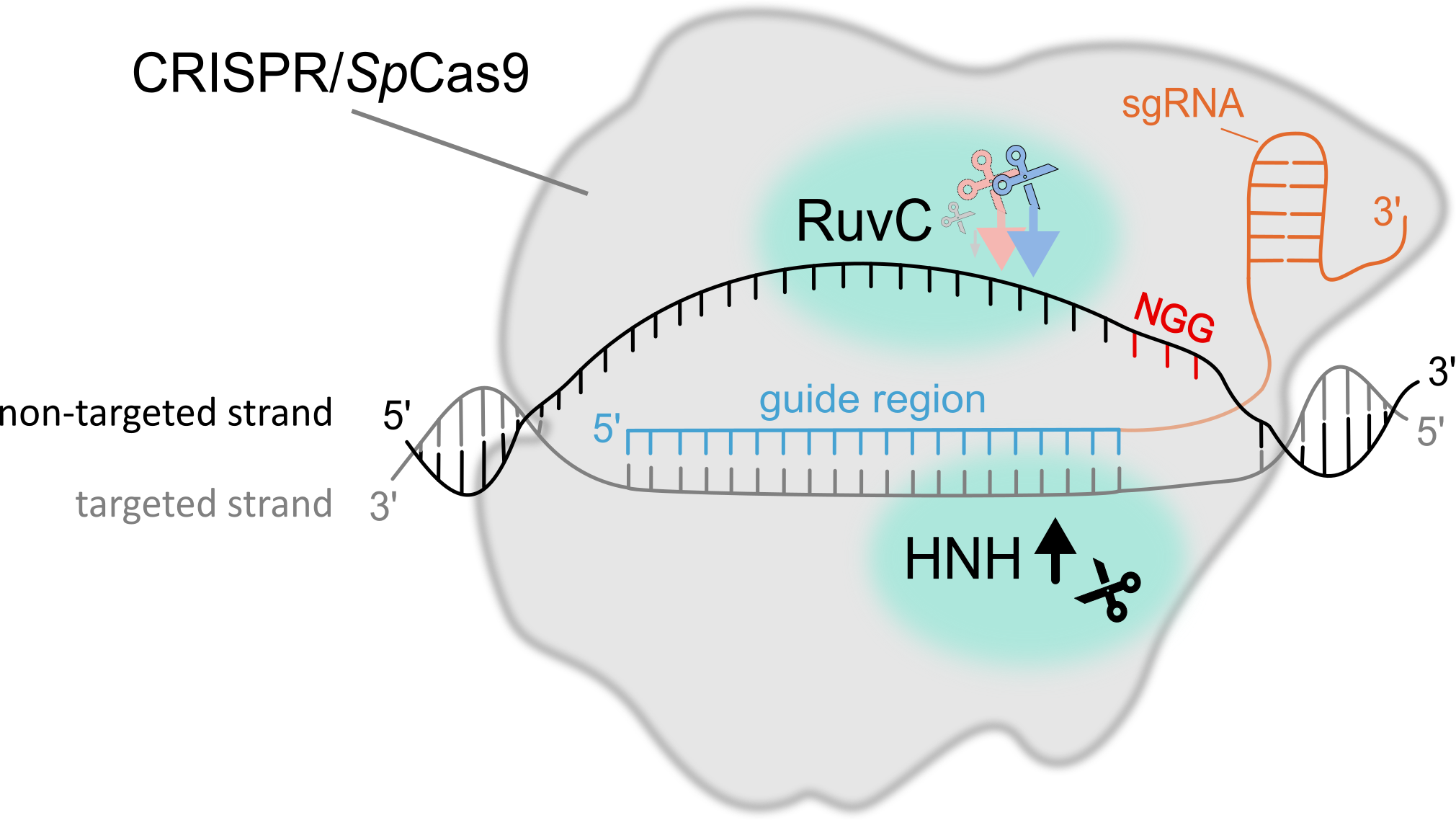
WHY ARE WE DOING THIS? CRISPR/Cas systems have become an indispensable tool in many fields of experimental biology, from basic research, where they are used, for example, to generate knock-outs of specific genes to study their function, to applied research and plant breeding. The demands on the precision of the technique and the speed of the process are high. It is, therefore, essential to know how the CRISPR/Cas mechanism works, what and how affects its binding to the target site, the type of cleavage and how the cleaved site is subsequently repaired, both at the level of individual cells and tissues. It is known that the efficiency at particular sites that are only tens to lower hundreds of nucleotides apart typically have very different efficiencies, even on the order of tens of per cent. Thus, more accurate prediction of CRISPR/Cas mutagenesis outputs can reduce the number of experiments required to achieve the desired results and thus significantly reduce the time required for experiments and overall costs.
WHAT WE ALREADY KNOW? Many of the details describing the various stages of the process, from locating the target site for editing to the resulting repair of the generated double-strand break, have already been well described. However, little is known about how the epigenome affects target site search, residence time at the target site, type of cleavage, and the subsequent repair of the resulting break. An up-to-date summary of how the CRISPR/Cas9 system works, what influences it and how to use it can be found in our review 'How to use CRISPR/Cas9 in plants: from target site selection to DNA repair'.
In our paper 'DNA methylation can alter CRISPR/Cas9 editing frequency and DNA repair outcome in a target-specific manner', we described how targeted siRNA production (and subsequent introduction of epigenetic marks on DNA) against the promoter and coding region of a reporter transgene affects the resulting mutagenesis. Our findings suggest that DNA methylation (an epigenetic mark) can indirectly affect Cas9 activity and subsequent DNA repair, likely through changes in the local chromatin structure.
WHAT NOW AND WHAT THEN? In the follow-up research, we are looking in more detail at the effect of ongoing transcription and the presence of siRNA on the frequency and type of mutation. We also investigate the cytoskeleton's involvement in individual DNA double-strand break repairs and their visualization.
WANT TO KNOW MORE, WANT TO JOIN US? We currently have several bachelor's, diploma and master's theses on the following topics. If you are interested in some topic or have any own ideas about a research topic, please get in touch with our team leader Lukáš Fischer or postdocs Adéla Přibylová and Vojta Čermák (by email or drop by our lab in Viničná 5, 3rd floor, door 210 at the end of the corridor on the left).