Mgr. Adéla Přibylová, Ph.D.

Adéla works in Lukáš Fischer's Laboratory of Molecular Biology of Plants, and her scope of interest is epigenetics and its influence on CRISPR/Cas mutagenesis and DNA repair.
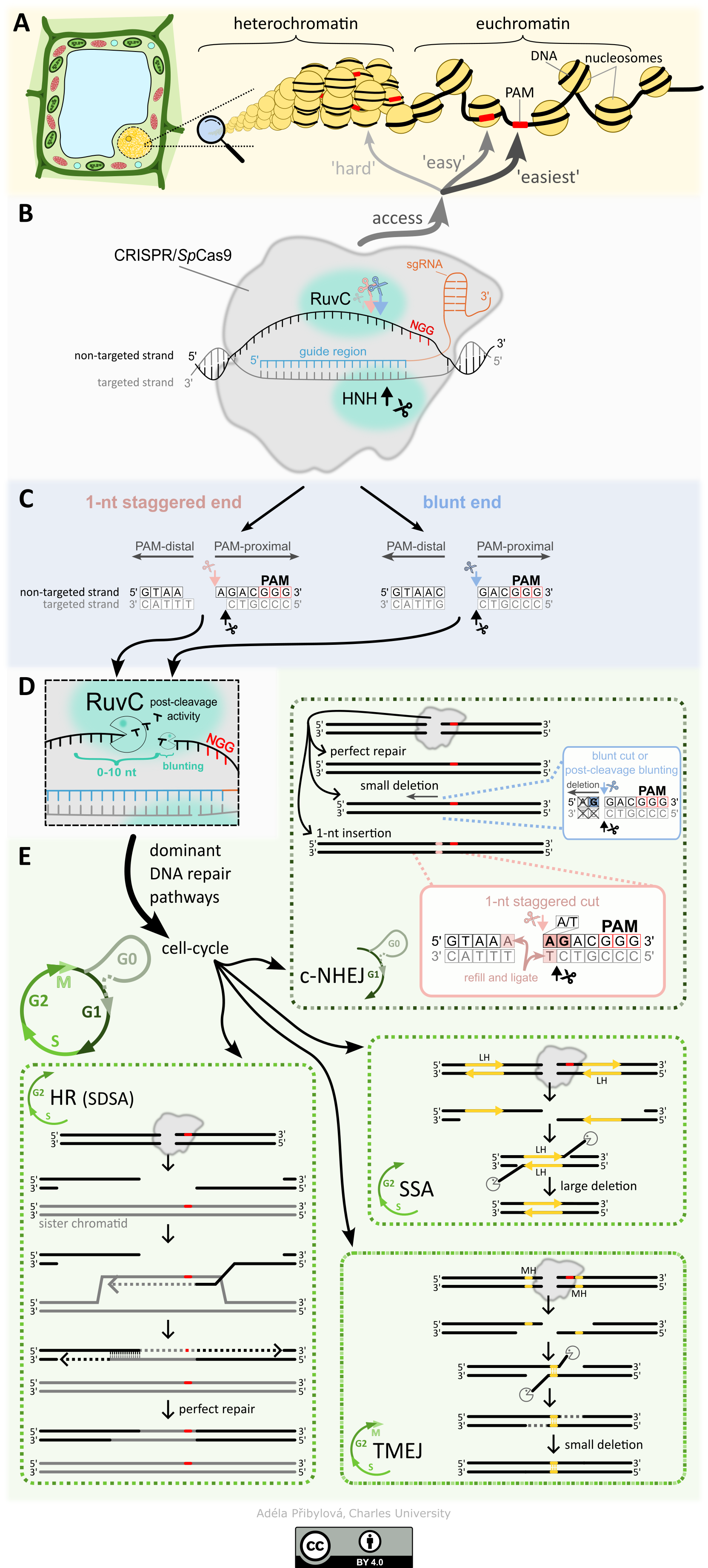
Figure description. Overview of CRISPR/SpCas9 mutagenesis in a plant cell. (A) The illustration shows two main chromatin states: euchromatin and heterochromatin. Euchromatin is more accessible to SpCas9 compared with heterochromatin. Similarly, SpCas9’s efficiency in locating the PAM sequence (red bar) is higher on the linker than on the nucleosome. (B) Schematic representation of CRISPR/SpCas9 bound in the target site. The targeted strand (grey) pairs with the guide region (blue) of the sgRNA (orange). The cleavage is facilitated by the endonuclease HNH and RuvC domains, which cut the targeted and non-targeted strands, respectively. (C) Depiction of two primary DNA end types following SpCas9 cleavage. The blunt end (blue, on the right) is cleaved by both endonuclease domains just 3 nt upstream of the PAM. The 1 nt staggered end (pink, on the left) is cleaved 3 nt upstream of the PAM on the targeted strand and 4 nt upstream of the PAM on the non-targeted strand. DNA ends containing the PAM are termed PAM-proximal, while those without are termed PAM-distal. (D) After cleavage, the non-targeted strand may undergo post-cleavage processing via the activity of the RuvC domain. PAM-distal ends can be trimmed by up to 10 nt, while PAM-proximal ends can be blunted if a staggered end has been created by the primary RuvC cut. The extent of the post-cleavage activity is determined by the duration for which SpCas9 remains bound at the target site. (E) Overview of the four main pathways participating in SpCas9-induced double-strand break (DSB) repair. The selection of the repair pathway in the cell is strongly influenced by the cell cycle stage. The dominant pathway in the G1 and G0 phases is the classical non-homologous end joining (c-NHEJ). It results in either a perfect repair, a small deletion (commonly on the PAM-distal side; occurs mainly when the fourth nucleotide upstream of the PAM is G), or a 1 nt insertion (mostly duplication of the fourth nucleotide upstream from PAM, especially when the fourth nucleotide is A or T and the third nucleotide is G). During the S and G2 phases, DSBs can be repaired by the homologous recombination (HR), single-strand annealing (SSA), or polymerase-theta-mediated end joining (TMEJ) pathways. HR requires a sister chromatid or other template (grey), the 5' strands of DNA ends are resected, and the exposed 3' tail invades into the sister chromatid serving as a template for the repair (the scheme shows HR subcategory synthesis-dependent strand annealing; SDSA: see ‘Homologous recombination’ for more details). After the tail is elongated according to the homologous template, the invading strand is displaced and pairs with its complementary strand from the opposite side of the DSB; both 3' ends are elongated to refill the remaining gaps, and the resulting nicks are ligated. HR usually results in a perfect repair. SSA repair is mediated through direct repeats (long homology; LH) around the DSB. Similar to HR, the 5' ends of the DSB are resected, and thereafter the 3' tails hybridize together via the LH region. Then, 3' flaps are trimmed, followed by ligation of the remaining nicks, leading to larger deletions. The TMEJ pathway uses short (mostly 2–6 nt) microhomologies (MH, yellow) occurring near the DSB. In addition to the S and G2 phases of the cell cycle, it also occurs in the M phase. In TMEJ, the 5' ends are shortly resected, Polθ identifies MHs on opposite sides of the DSB, the 3' flaps are removed, and the remaining gaps beyond the 3' ends of the MH are filled in, followed by ligation of individual nicks. TMEJ results in small deletions.
The scheme was published in work: Přibylová A, Fischer L. How to use CRISPR/Cas9 in plants: from target site selection to DNA repair. Journal of Experimental Botany, (2024) 10.1093/jxb/erae147
- ORCID ID:
- 0000-0001-8675-5065
- ResearcherID:
- H-5497-2017
- Scopus Author ID:
- 57210977726





















