It is well known that cats can take a number of shapes. It is also known that protein sequences fold in space into particular conformations. Like cats however, proteins too can change their shape depending on the situation, for example, in the presence of a binding partner.
So what happens to the structure of a protein when it binds a ligand? Moreover, what happens to the protein binding site; does it change, and if so, how or how much? Questions like these are very important for understanding the behaviour and dynamics of proteins, and are therefore relevant to drug design.
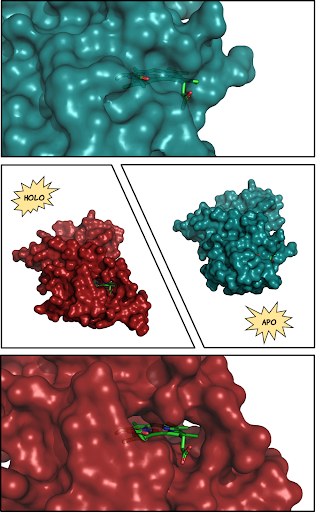
We developed a pipeline called AHoJ (Apo-Holo Juxtaposition), that finds and compares existing experimentally determined structures of proteins from the Protein Data Bank (PDB). We then used AHoJ to build a database by using protein binding sites as search queries (specified through their interacting ligand), to find existing structures in the PDB that feature this binding site, and group them together. Depending on whether they bind a ligand or not, the retrieved binding sites are annotated as holo or apo respectively.
A number of metrics are also collected to compare the different states of the binding sites (local and global RMSD, solvent accessible surface area, and more). In AHoJ-DB we look at ~500,000 binding sites and find that less than half have an apo form in the entire known structure space (PDB). Also, the apo states of the binding sites have five times more disordered residues on average, compared to the holo; that is, residues that are supposed to be in the structure, but are missing, possibly on account of increased mobility in the absence of their binding partner.
Interestingly, the availability of apo forms for binding sites of different ligands varies considerably: for the binding sites of the 15 most abundant ligands it ranges between 1 to 73%. In other words, binding sites that bind certain ligands are much more likely to have a known apo form, and may therefore be more ordered than others.
Furthermore, when aligning the apo and holo states onto each other, there is a significant fluctuation in both global (protein chain) and especially local (binding site) RMSD, showing that the conformation of proteins often changes between these two forms. Binding sites that show significant changes are known as cryptic binding sites and are particularly interesting in drug design.
Read more: https://doi.org/10.1016/j.jmb.2024.168545
Try AHoJ & AHoJ-DB: https://apoholo.cz/





















